Multalin result page






Go directly to Alignment
Multalin version 5.4.1
Copyright I.N.R.A. France 1989, 1991, 1994, 1996
Published research using this software should cite
Multiple sequence alignment with hierarchical clustering
F. CORPET, 1988, Nucl. Acids Res., 16 (22), 10881-10890
Symbol comparison table: blosum62
Gap weight: 12
Gap length weight: 2
Consensus levels: high=90% low=50%
Consensus symbols:
! is anyone of IV
$ is anyone of LM
% is anyone of FY
# is anyone of NDQEBZ
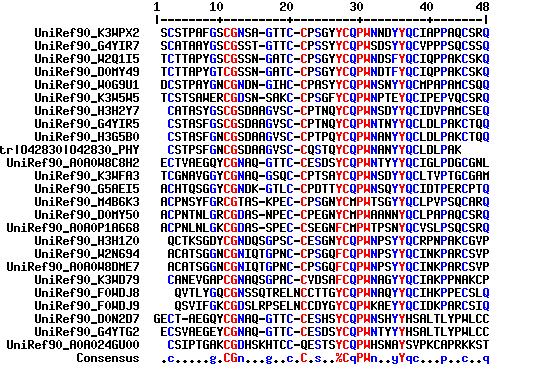
MSF: 48 Check: 0 ..
Name: UniRef90_K3WPX Len: 48 Check: 9611 Weight: 0.82
Name: UniRef90_G4YIR Len: 48 Check: 2519 Weight: 0.82
Name: UniRef90_W2Q1I Len: 48 Check: 9264 Weight: 0.47
Name: UniRef90_D0MY4 Len: 48 Check: 9308 Weight: 0.47
Name: UniRef90_W0G9U Len: 48 Check: 9533 Weight: 0.96
Name: UniRef90_K3W5W Len: 48 Check: 9963 Weight: 1.09
Name: UniRef90_H3H2Y Len: 48 Check: 9564 Weight: 0.72
Name: UniRef90_G4YIR Len: 48 Check: 86 Weight: 0.55
Name: UniRef90_H3G5B Len: 48 Check: 9459 Weight: 0.55
Name: tr|O42830|O428 Len: 48 Check: 3630 Weight: 0.98
Name: UniRef90_A0A0W Len: 48 Check: 8894 Weight: 1.17
Name: UniRef90_K3WFA Len: 48 Check: 9395 Weight: 1.17
Name: UniRef90_G5AEI Len: 48 Check: 417 Weight: 1.23
Name: UniRef90_M4B6K Len: 48 Check: 482 Weight: 1.00
Name: UniRef90_D0MY5 Len: 48 Check: 7810 Weight: 0.90
Name: UniRef90_A0A0P Len: 48 Check: 341 Weight: 0.90
Name: UniRef90_H3H1Z Len: 48 Check: 413 Weight: 1.17
Name: UniRef90_W2N69 Len: 48 Check: 356 Weight: 0.66
Name: UniRef90_A0A0W Len: 48 Check: 356 Weight: 0.66
Name: UniRef90_K3WD7 Len: 48 Check: 7298 Weight: 1.41
Name: UniRef90_F0WDJ Len: 48 Check: 796 Weight: 1.39
Name: UniRef90_F0WDJ Len: 48 Check: 9154 Weight: 1.39
Name: UniRef90_D0N2D Len: 48 Check: 11 Weight: 1.21
Name: UniRef90_G4YTG Len: 48 Check: 585 Weight: 1.21
Name: UniRef90_A0A02 Len: 48 Check: 508 Weight: 2.11
Name: Consensus Len: 48 Check: 1400 Weight: 0.00
//
Available files:
-Sequence Input file
-Cluster file
-Results as a fasta file
-Results as a text page (msf)
-Results as postscript page(s)
with ESPript (protein only  )
)
-Alignment and tree description (rfd)
Get a better view of your protein family : phylogenetic tree, pruned
tree and subtrees, summarised coloured alignment and subalignments.
-
Results as an html page (needs to enable style sheets)
-Results as a text page with colour indications
(need a text editor)
-Results as a gif image